Recent Publication: Method for Discovery of Structural Variations in Hard-to-Call Genome Regions
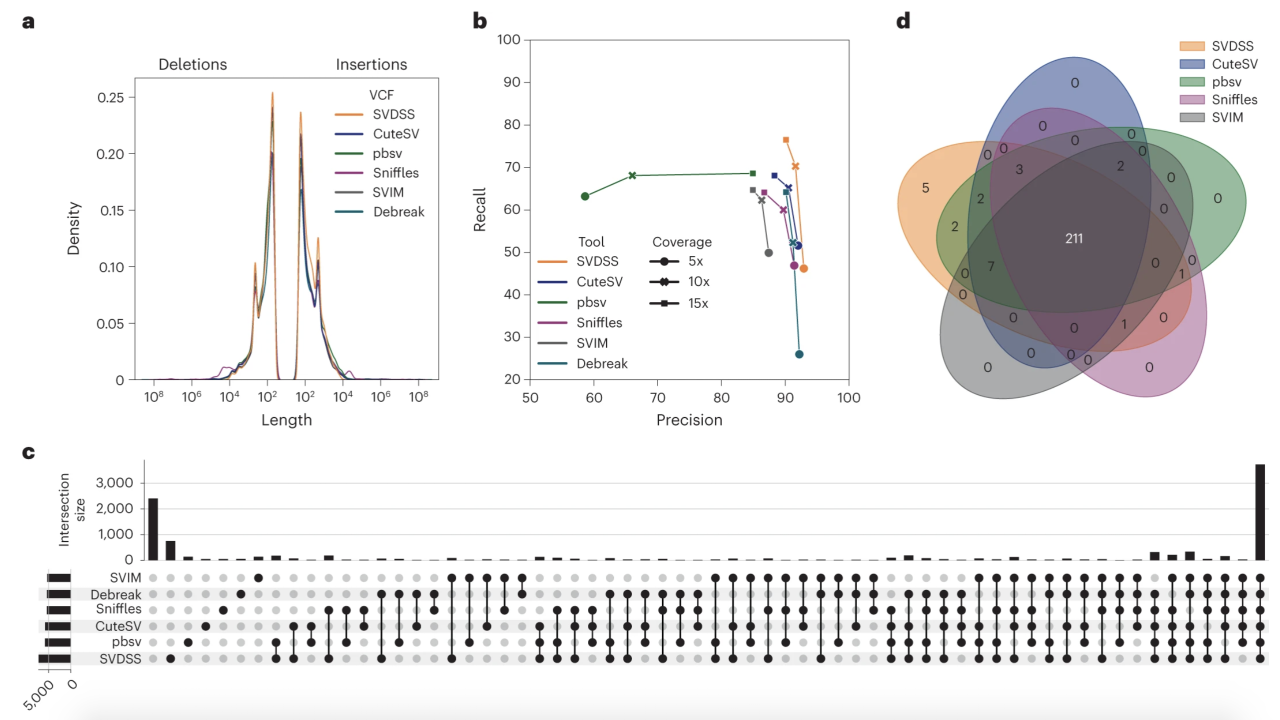
Published in Nature Methods in December 2022, the paper titled “SVDSS: structural variation discovery in hard-to-call genomic regions using sample-specific strings from accurate long reads” by Denti, Khorsand, Bonizzoni, Hormozdiari, and Chikhi describes a newly developed method to discover structural variants. Structural variants are defined as medium to large size genetic variations and are shown to be important in human genomics, evolution, and diseases. They are also the least well-characterized type of genetic variation. The proposed novel approach utilizes a concept previously introduced by the same team denoted as Sample-Specific Strings (Khorsand et al., 2021).
Studying structural variants that reside in genomic regions with repeats are challenging; however, this study presents a discovery method, SVDSS: SV Discovery with Sample-Specific strings, that uses long-read sequencing technologies and a combination of mapping-free, mapping-based, and assembly-based methods for improved discovery of structural variants. SVDSS is particularly effective at accurately predicting structural variants in repetitive and hard to genotype regions of the genome.
Media Resources
Denti L, Khorsand P, Bonizzoni P, Hormozdiari F, Chikhi R. 2022. SVDSS: structural variation discovery in hard-to-call genomic regions using sample-specific strings from accurate long reads. Nature Methods. DOI: 10.1038/s41592-022-01674-1.
Khorsand P, Denti L, Bonizzoni P, Chikhi R, Hormozdiari F. 2021. Comparative genome analysis using sample-specific string detection in accurate long reads. Bioinformatics Advances. DOI: 10.1093/bioadv/vbab005
