Bioinformatics Core
bioinformatics.core@ucdavis.edu
Core Manager: Jean Challacombe
Core Office: 530-752-2698
Set up a PPMS account here (instructions here)
 The Bioinformatics Core provides expertise and infrastructure for the acquisition, curation, analysis, and distribution of large complex biological datasets, with an emphasis on sequence data. We offer data analysis services, free consultations for grant or project planning, and training aimed at empowering graduate students, post-docs, and PIs to do their own bioinformatics data analysis. Also, we can help you design, install, and maintain your lab's high-performance computing infrastructure, providing stable support even as lab members come and go. Please send us an email or call (530-752-2698) with your questions, or visit our website to find out more about how we can help you.
The Bioinformatics Core provides expertise and infrastructure for the acquisition, curation, analysis, and distribution of large complex biological datasets, with an emphasis on sequence data. We offer data analysis services, free consultations for grant or project planning, and training aimed at empowering graduate students, post-docs, and PIs to do their own bioinformatics data analysis. Also, we can help you design, install, and maintain your lab's high-performance computing infrastructure, providing stable support even as lab members come and go. Please send us an email or call (530-752-2698) with your questions, or visit our website to find out more about how we can help you.
DNA Technologies & Expression Analysis Core
Core Manager: Dr. Lutz Froenicke
Lab: 530-754-9143 / Fax: 530-754-9658
Set up a PPMS account here (instructions here)
 The DNA Technologies and Expression Analysis Core offers high-throughput and custom sequencing, genotyping, and microarray services, as well as trainings and consultations. The lab is a designated UC Davis Campus Research Core Facility.
The DNA Technologies and Expression Analysis Core offers high-throughput and custom sequencing, genotyping, and microarray services, as well as trainings and consultations. The lab is a designated UC Davis Campus Research Core Facility.
The core offers the three complementary Next Generation Sequencing (NGS) technologies: Illumina sequencing, and PacBio and Nanopore (long read) sequencing, providing the full spectrum of sequencing options and a wide range of library preparation services for all platforms. Genotyping is performed by sequencing (targeted sequencing and reduced-representation sequencing) as well as on the Fluidigm EP1 and on Illumina Infinium arrays. Gene expression analysis is carried out by RNA-seq on HiSeq, NextSeq, and NovaSeq sequencers. Single-cell transcriptome (high-throughput single-cell gene expression profiling) and genome analyses (linked-read whole genome sequencing) are enabled by our 10X Genomics Chromium System. The core offers annual Illumina and PacBio sequencing library preparation workshops, free consultations on project considerations and experimental design, and custom sequencing library prep solutions. Many of the core’s instruments are available to scientists on campus as shared equipment for a small usage fee after training. We operate a PacBio Sequel, Oxford Nanopore PromethION, Illumina HiSeq 4000, NextSeq, and MiSeq sequencers, and sequence on an Illumina NovaSeq 6000. The sequencers are complemented by liquid handling robots (PerkinElmer’s Sciclone NGS G3, IntegenX’s Apollo, Opentrons OT2), a Covaris E220 sonicator, and a Fluidigm Access Array system for high-throughput sample and sequencing library preparation. The liquid handling robots are employed to minimize sample handling variation and to provide fast turnaround times. The core maintains a data portal allowing users fast access to the data. We offer joint consultations with the neighboring Bioinformatics Core, which provides sequence data analysis and statistical evaluations. Gene expression studies and high-quality de novo genome assemblies are two of the focal points of the Cores.
West Coast Metabolomics Center (Metabolomics Core)
WCMC Technical Director: Dr. Uri Keshet
Lab: 530-754-8553 / Fax: 530-754-8370
Set up a PPMS account here (instructions here)

The Metabolomics Core provides untargeted and metabolite target analyses for a wide range of biochemical pathways, covering over 1,200 identified metabolites with validated analytical methods and known mass spectra and retention times. The facility utilizes eight LC-MS and seven GC-MS instruments with both nominal mass and accurate mass capabilities. The core is happy to engage in project-specific method development and implementation.
The core further provides services in univariate and multivariate statistical analysis of data sets, pathway analysis and mapping, partial correlation, and network analysis. We are looking forward to collaborating with you for turning your metabolomic data into biologically significant discoveries!
Proteomics Core
Core Manager: Dr. Brett Phinney
Lab: 530-754-5298 / Fax: 530-754-8370
Set up a PPMS account here (instructions here)
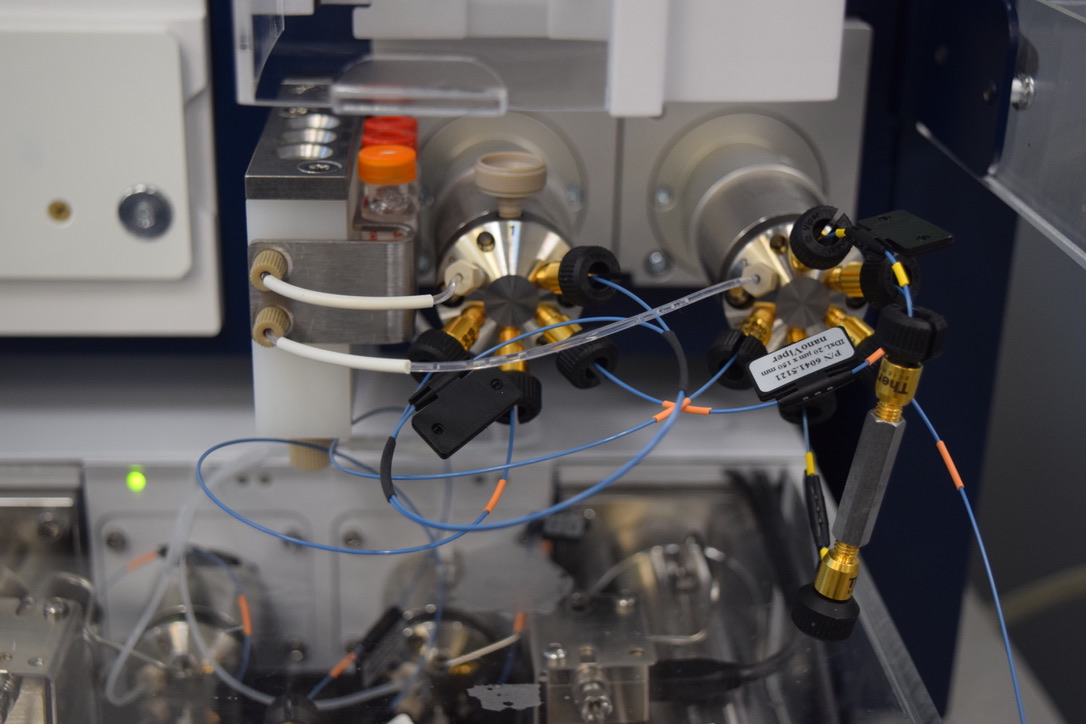 The Proteomics Core provides state-of-the-art analytical proteomic services with particular emphasis on label free quantitative proteomic profiling, the analysis of macromolecular complexes, the post-translational modification of their constituents and standard protein identification from complex protein mixtures.
The Proteomics Core provides state-of-the-art analytical proteomic services with particular emphasis on label free quantitative proteomic profiling, the analysis of macromolecular complexes, the post-translational modification of their constituents and standard protein identification from complex protein mixtures.
The core has a staff of six individuals including two Ph.D. scientists. For high throughput identification and characterization of proteins and peptides the core has at its disposal several state of the art LC-MS/MS systems including a new Thermo Fisher Scientific Q-Exactive Orbitrap mass spectrometer connected to an EASY nLC II system, a LTQ-FT Ultra hybrid Fourier Transform Ion Cyclotron tandem mass spectrometer connected to a Waters Acuity Ultra High Pressure Nano UPLC, two Thermo Scientific LTQ tandem mass spectrometers connected to a Michrom Paradigm HPLC’s, and a Thermo Scientific TSQ Vantage Triple Quadrupole mass spectrometer for quantitative multiple reaction monitoring (MRM) targeted proteomics experiments. Core personnel can also provide amino acid analysis using one of three Hitachi amino acid analyzers and N-terminal sequencing using one of two N-terminal sequencers if needed.
TILLING Core
Core Manager: Dr. Luca Comai
Lab: 530-754-1316
Website here
Set up a PPMS account here (instructions here)
 The TILLING Core provides mutants for rice, Arabidopsis thaliana, and tomato as well as other species on request. TILLING (Targeting Induced Local Lesions IN Genomes) is a reverse genetic technique that uses chemical mutagenesis to create libraries of mutagenized individuals that are subjected to high-throughput sequencing for the discovery of mutations.
The TILLING Core provides mutants for rice, Arabidopsis thaliana, and tomato as well as other species on request. TILLING (Targeting Induced Local Lesions IN Genomes) is a reverse genetic technique that uses chemical mutagenesis to create libraries of mutagenized individuals that are subjected to high-throughput sequencing for the discovery of mutations.
Kitbase
Complementary to the TILLING Core is Kitbase, a public resource and database for KitaakeX mutants.

